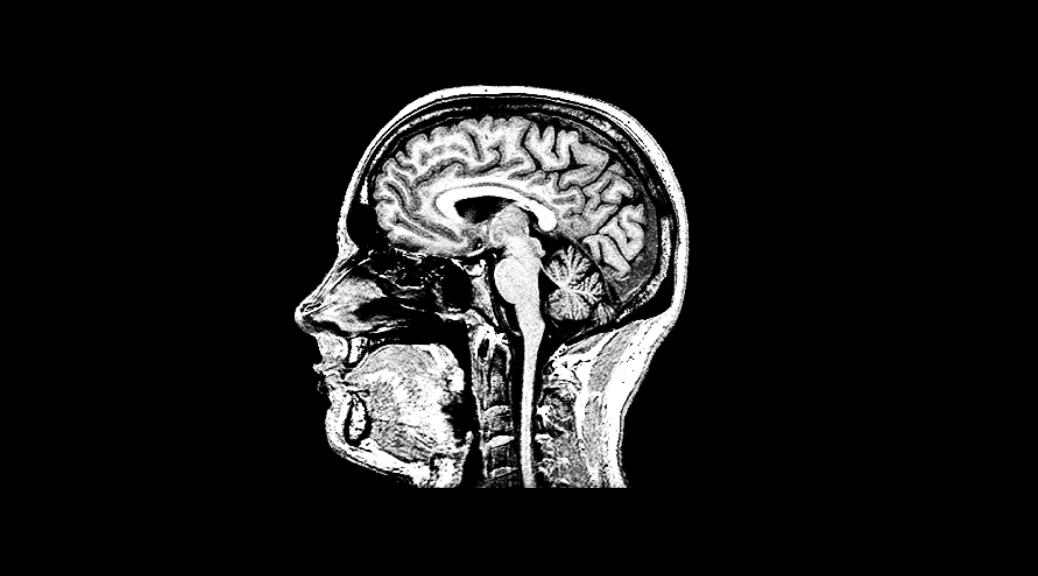
If you get an MRI scan done, they provide you a CD with the images. However, these images are in a binary format and can only be viewed using the Windows based software on the CD. What do you do if you have a Mac or a Linux PC? OR if you want to share the images online? Here is a Python script that can help.
I’ve had two of my family members reach out to me for help with doing this. They had scans and they wanted to be able to share them across the internet with another doctor. Sending them the files inside the CD didn’t help much and the patient wasn’t sure how to use the CD themselves.
For the first time, I spent hours opening each image in the software on the CD and using screen capturing capabilities to convert each binary image to a standard JPEG image. It was a long laborious process but I got the job done.
Recently, I got a second request for a family member that had close to a 100 scans. He had all Apple computers so he couldn’t even view the scan him selves, let alone send them to anyone! This time, I decided to look for a programatic solution to be able to batch convert these files.
Python came to the rescue. I was able to put together a conversion script in an hour using the pydicom package to read the binary files and use the pypng package to write the converted images.
You can find the script at Github here OR check out the code below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 |
import os import png import dicom def mri_to_png(mri_file_path, png_file_path): """ Function to convert an MRI binary file to a PNG image file. @param mri_file_path: Full path to the mri file @param png_file_path: Fill path to the png file """ # Extracting data from the mri file plan = dicom.read_file(mri_file_path) shape = plan.pixel_array.shape image_2d = [] max_val = 0 for row in plan.pixel_array: pixels = [] for col in row: pixels.append(col) if col > max_val: max_val = col image_2d.append(pixels) # Rescalling greyscale between 0-255 image_2d_scaled = [] for row in image_2d: row_scaled = [] for col in row: col_scaled = int((float(col)/float(max_val))*255.0) row_scaled.append(col_scaled) image_2d_scaled.append(row_scaled) # Writing the PNG file f = open(png_file_path, 'wb') w = png.Writer(shape[0], shape[1], greyscale=True) w.write(f, image_2d_scaled) f.close() def convert_folder(mri_folder, png_folder): """ Convert all MRI files in a folder to png files in a destinaiton folder """ os.makedirs(png_folder) for mri_file in os.listdir(mri_folder): mri_file_path = os.path.join(mri_folder, mri_file) png_file_path = os.path.join(png_folder, '%s.png' % mri_file) try: mri_to_png(mri_file_path, png_file_path) print mri_file_path, '-->', png_file_path except: print 'FAIL>', mri_file_path, '-->', png_file_path |
I am hoping to convert this capability into a web servive that can potentially help a lot of people. The only issues I see are related to patient data confidentiality. I’m going to start looking into this, if someone has any suggestions or thoughts, I’d love to hear.